Previous issue | Next issue | Archive
Volume 10 (3); May 25, 2020![]() [Booklet]
[Booklet]
COVID-19 and avian corona viruses: epidemiological comparison and genetic approach
| COVID-19 and avian corona viruses: epidemiological comparison and genetic approach |
Berghiche A.
J. Life Sci. Biomed., 10(3): 21-28, 2020; pii:S225199392000004-10
DOI: https://dx.doi.org/10.51145/jlsb.2020.4
Abstract
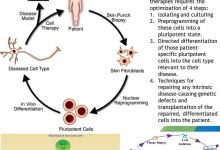
Aim. This study aimed to collect and analyse available information on COVID-19 and avian corona viruses in order to conduct a systematic review of the genetic data concerning them. Methods. All available research was done according to the strictest data selection criteria, and the databases like NCBI genebank were quantitatively searched in the currently available scientific literature using keywords, analytical statistic and genomic software. All studies on the coronavirus family were dedicated to provide an overview towards an advanced statistical analysis of the collected data. The first step was a descriptive study of COVID-19 and avian corona viruses by an epidemiological comparison between the two cases. Results. All corona viruses usually tend to have relatively A-T rich DNAs which is linked to their highly A-T rich codon biases. The results indicate genetic differences between the two viruses, but the results of a percentage analysis showed that the nucleotides A+T are both more abundant and energetically cheaper than nucleotides G+C, this gives viruses a selection advantage. Conclusion. These results give us a future positive view of this type of virus with AT-rich genomes which is selectively promoted at the host level. Recommendation. A recommendation by our study reveals that thought about the vaccine is very early but prevention has proven to be effective for this virus in chickens.
Keywords: Avian corona virus, COVID-19, Epidemiological comparison, Genomic analysis.
[Full text-PDF] [HTML] [ePub] [XML] [Export citation as BibTeX, RIS & EndNote] [How to Cite] [Google Scholar] [Dimensions ID]
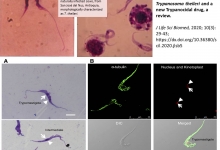
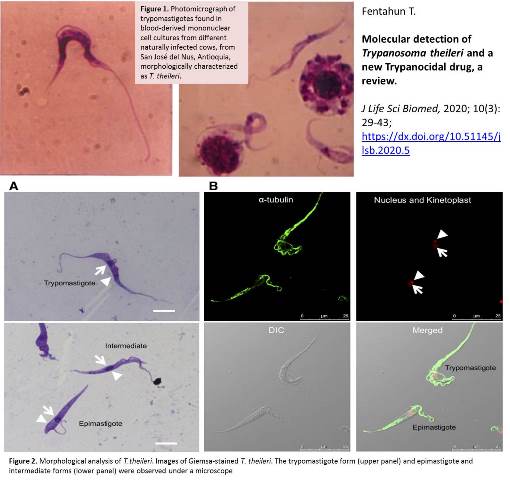
Molecular detection of Trypanosoma theileri and a new Trypanocidal drug, a review
| Molecular detection of Trypanosoma theileri and a new Trypanocidal drug, a review |
Fentahun T.
J. Life Sci. Biomed., 10(3): 29-43, 2020; pii:S225199392000005-10
DOI: https://dx.doi.org/10.51145/jlsb.2020.5
Abstract
Introduction. Trypanosoma theileri (T.theileri) is apathogenic, cosmopolitan and commensal protozoa of cattle. Despite apathogenic in healthy, but not in stressed cattle; it’s getting recent attention as a tool to tackle pathogenic ones. These days, researchers are giving due attention to study the biology and feasibility of T.theilerito use it as a model candidate for novel drug discovery. In addition, in-silico analysis using common antitrypanosome drug targets couldn’t show significant similarity both at DNA and protein level. Nevertheless, homologous sequences have been identified among drug targets for Ornithine decarboxylase. This indicated the possibility to consider T.theileri as a model to search novel drugs once having whole genome sequences. The SDM 79 is an appropriate medium to cultivate at 26 oC, without CO2. Gradient PCR amplification has been used to detect using T. theileri. The specific primer (Tth625) which reveals 465 bp amplification product and also the full length 18S ribosomal DNA sequence of T.theileri DNA detectable at 730 base pairs are commonly used. Whole genome and transcriptome analysis can show the phylogenetic relationship between T.theileri and other pathogenic Trypanosomes which can be the basis for novel drugs development. Aim. The purpose of this paper is toreview the nature of Trypanosoma theileri, its molecular identification and also using its apathogenic nature as an opportunity to discover anew Trypanocidal drug.
Keywords: Model, Novel Trypanocidal drug, PCR, SDM 79, Trypanosoma theileri
[Full text-PDF] [HTML] [ePub] [XML] [Export citation as BibTeX, RIS & EndNote] [How to Cite] [Semantic Scholar] [Dimensions ID]