Previous issue | Next issue | Archive
| Phylogenetic analysis and antibacterial potency of indigenous mushrooms in Arabuko Sokoke forest, Kenya |
Research Paper
Phylogenetic analysis and antibacterial potency of indigenous mushrooms in Arabuko Sokoke forest, Kenya
Audrey MS, Kamau PK, and Neondo J. J. Life Sci. Biomed., 15(4): 78-102, 2025; pii:S225199392500010-15
J. Life Sci. Biomed., 15(4): 78-102, 2025; pii:S225199392500010-15
DOI: https://dx.doi.org/10.54203/jlsb.2025.10
Abstract
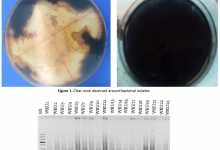
Indigenous mushrooms are a rich source of bioactive compounds with potential pharmacological application. This study investigated the diversity, molecular identity and antibacterial activity of indigenous mushroom collected from Arabuko Sokoke forest, Kenya. Fifteen mushroom accessions were purposively sampled and subjected to DNA extraction using an optimized CTAB protocol. Molecular identification was carried out through ITS1 and ITS4 amplification followed by sequencing and phylogenetic analysis. Crude extracts of the mushrooms were screened for antibacterial activity against Escherichia coli (Gram-negative) and Staphylococcus aureus (Gram-positive) using the disc diffusion method. Active extracts were further profiled using Gas Chromatography-Mass Spectrometry to identify bioactive metabolites. Molecular docking was performed to predict compound-protein interactions with bacterial targets. Sequencing and phylogenetic analysis identified 12 distinct taxa, including Ganoderma mbrekobenum, Leiotrametes lactinea, Trametes polyzona and Auricularia polytricha. Among the 15 extracts, only G. mbrekobenum (MU005) and L. lactinea (MU012) exhibited antibacterial activity. The two mushrooms produced zones of inhibition of 24.1-34.3mm, which were comparable to or greater than ampicillin controls. Gas Chromatography- Mass Spectrometry profiling of these two mushrooms revealed four bioactive compounds: 2,4-di-tert-butylphenol, 3-deoxy-D-mannoic lactone, 4-thiazolecarboxylic acid derivative, and 2-formylamino-3-methylbut-2-enoic acid (unique to G. mbrekobenum). Molecular docking indicated strong binding affinities of these compounds for key bacterial enzymes, including dihydrofolate reductase, enolpyruvyl transferase, phospholipase A2, and penicillin-binding protein 1, which supports the observed antibacterial activity. The findings highlight G. mbrekobenum and L. lactinea as promising sources of antibacterial agents, with activity comparable to standard antibiotics. The integration of molecular identification, chemical profiling, and in silico docking provides a robust framework for further discovery of natural products. Further studies should expand antibacterial screening to more pathogens, determine minimum inhibitory concentration and minimum bactericidal concentration values, and validate compound activity through bioassay-guided fractionation.
Keywords: Antibacterial activity, Ganoderma mbrekobenum, GC-MS, Leiotrametes lactinea, Molecular docking.
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
| Microbial screening for antimicrobial residues in cattle tissues processed at slaughterhouses in Kilosa District, Morogoro, Tanzania |
Research Paper
Microbial screening for antimicrobial residues in cattle tissues processed at slaughterhouses in Kilosa District, Morogoro, Tanzania
Mgonja FR, Mpina MC, Mwega ED, Temba BA, Shabaan M, and Mwangwengwa LM.
J. Life Sci. Biomed., 15(4): , 2025; pii:S225199392500011-15
DOI: https://dx.doi.org/10.54203/jlsb.2025.11
Abstract
Antimicrobial residues refer to drug molecules that remain in the meat and organs of slaughtered animals previously treated with antibiotics, particularly when withdrawal periods are not observed. The inappropriate use of antimicrobial drugs, limited knowledge among livestock stakeholders, and practices such as self-medication contribute to the accumulation of these residues in edible tissues. This study aimed to investigate the presence of antimicrobial residues in cattle tissues using microbial screening methods at slaughterhouses in Kilosa District. A cross-sectional study was conducted in three wards: Kimamba, Dumila, and Kilosa. A total of 90 tissue samples 30 each of muscle, liver, and kidney were collected. The samples were packaged in sterile polystyrene bags and transported to the laboratory at Sokoine University of Agriculture for analysis. Microbial inhibition methods were used to screen for the presence of antimicrobial residues. Data were analyzed using SPSS version 25. The chi-square test was employed to assess differences among tissue types, while Fisher’s exact test was used to evaluate associations. Overall, 28.9% (26/90) of the tissue samples tested positive for antimicrobial residues. Liver tissues showed the highest prevalence at 46.7% (14/30), followed by muscle tissues at 40.0% (12/30). Surprisingly, no antimicrobial residues were detected in kidney samples. The differences in residue occurrence among the tissue types were statistically significant (p = 0.000). Residue occurrence in muscle tissues varied significantly by ward (p = 0.000), indicating a strong geographic influence. However, the distribution of residues in liver tissues did not show a statistically significant variation among wards (p = 0.314), suggesting a more uniform presence. These findings highlight the urgent need to strengthen regulatory frameworks, promote responsible antibiotic use in livestock production, and enhance surveillance systems. Addressing these issues is critical to protecting public health and combating the growing threat of antimicrobial resistance.
Keywords: Microbial screening methods, antimicrobial residues, antimicrobial, Kilosa district
[Full text-PDF-Inpress] [ePub] [Export citation from ePrint] [How to Cite]
Previous issue | Next issue | Archive
![]() This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)