Previous issue | Next issue | Archive
![]() Volume 14 (3); September 25, 2024
Volume 14 (3); September 25, 2024
| Anti-methanogenic effect of pyrogallol in Spirulina platensis – molecular docking and dynamics simulation on methyl-coenzyme M reductases |
Research Paper
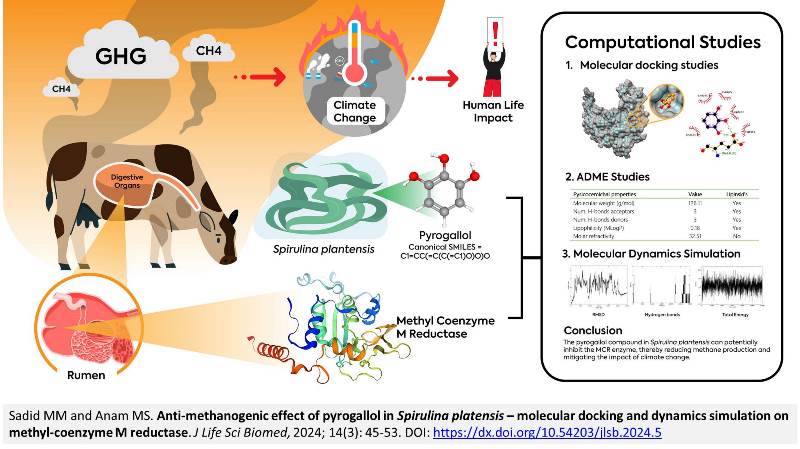
Anti-methanogenic effect of pyrogallol in Spirulina platensis – molecular docking and dynamics simulation on methyl-coenzyme M reductase
Sadid MM and Anam MS.
J. Life Sci. Biomed., 14 (3): 45-53, 2024; pii:S225199392400005-14
 DOI: https://dx.doi.org/10.54203/jlsb.2024.5
DOI: https://dx.doi.org/10.54203/jlsb.2024.5
Abstract
Methane, along with carbon dioxide and nitrogen oxides, is a key greenhouse gas contributing significantly to the global concern over climate change. This study investigated the anti-methanogenic properties of pyrogallol in Spirulina platensis using molecular docking and dynamics simulation on methyl-coenzyme M reductase (MCR). The Swiss ADME web server was used to identify pyrogallol's absorption, distribution, metabolism, and excretion (ADME) properties. Molecular docking studies were conducted using UCSF Chimera with the Vina script as the executor. The docking results were further analyzed through molecular dynamics simulation using Gromacs-2024. ADME analysis indicated that pyrogallol meets Lipinski’s Rule of Five. Docking studies revealed that pyrogallol has a binding affinity of 4.6 kJ/mol with 2 hydrogen bonds and 1 hydrophobic interaction. Additionally, the MCR-pyrogallol simulation results showed fluctuating root mean square deviation (RMSD) values that stabilized at t = 26,200 until the end of the simulation with an average value of 2.50 nm. Moreover, the hydrogen bonds formed during the simulation fluctuated, with no bonds observed for more than 75% of the simulation time. The energy released during the simulation reached –300.24 kJ/mol with an average of –5.19 kJ/mol. In conclusion, the pyrogallol compound in Spirulina plantensis can potentially inhibit the MCR enzyme, thereby reducing methane production and mitigating the impact of climate change.
Keywords: Anti-methanogenic, methyl-coenzyme M reductase, molecular docking, pyrogallol, Spirulina platensis
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
| Current vaccine candidate of toxoplasmosis |
Review
Current vaccine candidate of toxoplasmosis
Woldegerima E, Getachew F, Misganaw M, Mesfin Y, Belete D, Sisay T, Berhane N.
J. Life Sci. Biomed., 14 (3): 54-67, 2024; pii:S225199392400006-14
 DOI: https://dx.doi.org/10.54203/jlsb.2024.6
DOI: https://dx.doi.org/10.54203/jlsb.2024.6
Abstract
Toxoplasma gondii is an intracellular protozoan parasite belonging from the phylum Apicomplexa, known for causing toxoplasmosis. The disease has a global presence, affecting about one-third of the world’s population. The parasite infects various intermediate hosts, including humans and other warm-blooded mammals, with cats serving as the definitive hosts. Its life cycle is complex, featuring a sexual phase in the definitive host and an asexual phase in intermediate hosts. Toxoplasmosis can leads severe neurologic, ocular, and systemic diseases in neonates and immunocompromised individuals. In immunocompetent individuals, the infection is typically asymptomatic, forming dormant tissue cysts in immune-privileged sites such as the muscles and brain. During pregnancy, toxoplasmosis poses significant health risks, potentially causing severe birth defects or miscarriage, and a major concern for immunocompromised hosts. Current control measures are inadequate, highlighting the need for effective vaccines. The initial host defense against T. gondii occurs at the intestinal mucosa, where cytokines and chemokines released by intestinal epithelial cells facilitate the migration of inflammatory cells, including macrophages, neutrophils, and dendritic cells. Developing a vaccine that can enhance this mucosal immunity is crucial for preventing toxoplasmosis. Therefore, the development of vaccines against T.gondii is a promising alternative mechanism to prevent toxoplasmosis. This review aims to present the current status of vaccine candidates against Toxoplasma gondii.
Keywords: Vaccine, Candidate, Toxoplasmosis, Toxoplasma gondii
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
| Maternal outcomes in pregnant women with COVID-19: impact of disease severity, timing of intervention, and SARS-CoV-2 variants |
Research Paper
Maternal outcomes in pregnant women with COVID-19: impact of disease severity, timing of intervention, and SARS-CoV-2 variants
Alimova KP, Ibadov RA, Tyan TV, and Ibragimov SK.
J. Life Sci. Biomed., 14 (3): 68-76, 2024; pii:S225199392400007-14
 DOI: https://dx.doi.org/10.54203/jlsb.2024.7
DOI: https://dx.doi.org/10.54203/jlsb.2024.7
Abstract
This study aimed to analyze maternal outcomes in pregnant women infected with COVID-19, focusing on the impact of disease severity, timing of medical intervention, and the effect of different SARS-CoV-2 variants. A retrospective analysis was conducted on 9,288 pregnant women diagnosed with COVID-19. Disease severity was classified as mild (30%), moderate (52%), severe (17%), and critical (2%). The study also examined the timing of medical care, with 41.6% of women seeking care within 7 days of symptom onset and 58.4% after 7 days. The average gestational age was 271.8 days, with the majority (93.8%) at full-term pregnancy. Results indicated that 88.2% of women continued their pregnancies during treatment, while 11.9% experienced pregnancy termination, including preterm deliveries and miscarriages. The study found a significant (p<0.01) association between COVID-19 severity and adverse outcomes, such as preterm birth and maternal mortality. Severe and critical cases demonstrated increased risks of uteroplacental insufficiency (52.1% and 85.5%, respectively), the need for intensive respiratory support, and ICU admission. The differences in outcomes between the Delta and Omicron variants are also highlighted. The Delta variant was associated with more severe disease and higher rates of complications, including a higher need for cesarean sections, compared to the Omicron variant. The overall 30-day survival rate was 98.05%, with a noticeable drop to 43.59% in critically ill patients. This study underscores the importance of early medical intervention and continuous monitoring in managing COVID-19 in pregnant women. The findings also emphasize the need to consider the variant-specific effects of SARS-CoV-2 on maternal and neonatal outcomes, which can guide clinical decision-making and improve the prognosis for both mother and child.
Keywords: COVID-19, pregnancy, maternal outcomes, SARS-CoV-2 variants, preterm birth, uteroplacental insufficiency, respiratory support, intensive care, maternal outcomes
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
| Resistance gene detection database for antimicrobial resistance investigations emphasizing on genomics and metagenomics techniques |
Review
Resistance gene detection database for antimicrobial resistance investigations emphasizing on genomics and metagenomics techniques
Alemnew M, Gelaw A, Nibret K, Getu A, and Berhane N.
J. Life Sci. Biomed., 14 (3): 77-86, 2024; pii:S225199392400008-14
 DOI: https://dx.doi.org/10.54203/jlsb.2024.8
DOI: https://dx.doi.org/10.54203/jlsb.2024.8
Abstract
Antimicrobial resistance poses a grave threat to global health where bacteria become resistant to antimicrobials, rendering them ineffective against infections. It leads to increased illness, death, and healthcare costs. The overuse and inappropriate use of antibiotics in both human medicine and animal agriculture are the primary drivers of antimicrobial resistance. Methods for identifying antimicrobial resistance genes include culturing bacteria with antimicrobial susceptibility test, polymerase chain reaction, and whole genome sequencing for genomics and Metagenomics samples. Newer methods like whole genome sequencing are faster and more accurate. Metagenomics is a powerful tool that can be used to study antimicrobial resistance in various environments. It can study culturable and non-culturable bacteria and used to study samples from humans, animals, and the environment. Resistance gene detection databases serves as a centralized repository of knowledge about resistance genes, mechanisms, and trends of antimicrobial. Databases categorize resistance information by genetic factors, mechanisms, specific drugs, and drug families. This review focuses on powerful and updated databases for detecting resistance genes, including: CARD, ResFinder with pointFinder, ResFinderFG v2.0, MEGARes v3.0 and NDARO. This review aims to examine the significance of antimicrobial resistance databases and techniques in combating antimicrobial resistance. It compares the advantages and disadvantages of different databases for storing and techniques for identifying antimicrobial resistance genes. Additionally, it inform researchers in evaluating antimicrobial resistance study methodologies and database choices based on antimicrobial resistance factors such as microorganism type, study setting, data type, resistance gene nature, resistance focus and novelty of resistance mechanisms. The primary aim of this review is to compare different powerful databases and techniques for identifying ARGs, an issue that hasn't been thoroughly covered in other reviews. These databases provide valuable resources for researchers studying antimicrobial resistance, offering a comprehensive collection of resistance gene sequences and annotations. This knowledge is essential for developing innovative strategies to combat AMR and ensure the ongoing effectiveness of antibiotics.
Keywords: Antimicrobial Resistance, Database, Metagenomics, Resistance, Resistance gène
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
Previous issue | Next issue | Archive
![]() This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)