Previous issue | Next issue | Archive
![]() Volume 14 (4); December 25, 2024
Volume 14 (4); December 25, 2024
| An understanding of the latest pathophysiological mechanisms of pancreatic β-cells in type 2 diabetes |
Review
A
A
J. Life Sci. Biomed., 14(2): , 2024; pii:S225199392400004-13
DOI: https://dx.doi.org/10.54203/jlsb.2023.4
Abstract
O
Keywords:
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
| Therapeutic aspects of dietary fibre and glycemic index: a brief review |
Research Paper
P
D
J. Life Sci. Biomed., 14(2): 35-41, 2024; pii:S225199392400005-13
DOI:
Abstract
T
Keywords:
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
Previous issue | Next issue | Archive
![]() This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
Previous issue | Next issue | Archive
| Phylogenetic analysis and antibacterial potency of indigenous mushrooms in Arabuko Sokoke forest, Kenya |
Research Paper
Phylogenetic analysis and antibacterial potency of indigenous mushrooms in Arabuko Sokoke forest, Kenya
Audrey MS, Kamau PK, and Neondo J.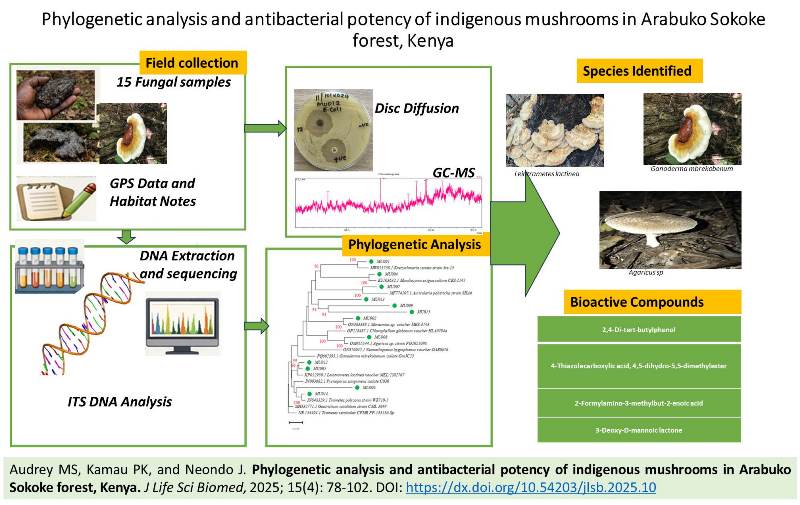 J. Life Sci. Biomed., 15(4): 78-102, 2025; pii:S225199392500010-15
J. Life Sci. Biomed., 15(4): 78-102, 2025; pii:S225199392500010-15
DOI: https://dx.doi.org/10.54203/jlsb.2025.10
Abstract
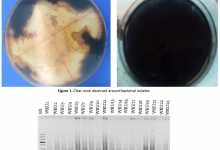
Indigenous mushrooms are a rich source of bioactive compounds with potential pharmacological application. This study investigated the diversity, molecular identity and antibacterial activity of indigenous mushroom collected from Arabuko Sokoke forest, Kenya. Fifteen mushroom accessions were purposively sampled and subjected to DNA extraction using an optimized CTAB protocol. Molecular identification was carried out through ITS1 and ITS4 amplification followed by sequencing and phylogenetic analysis. Crude extracts of the mushrooms were screened for antibacterial activity against Escherichia coli (Gram-negative) and Staphylococcus aureus (Gram-positive) using the disc diffusion method. Active extracts were further profiled using Gas Chromatography-Mass Spectrometry to identify bioactive metabolites. Molecular docking was performed to predict compound-protein interactions with bacterial targets. Sequencing and phylogenetic analysis identified 12 distinct taxa, including Ganoderma mbrekobenum, Leiotrametes lactinea, Trametes polyzona and Auricularia polytricha. Among the 15 extracts, only G. mbrekobenum (MU005) and L. lactinea (MU012) exhibited antibacterial activity. The two mushrooms produced zones of inhibition of 24.1-34.3mm, which were comparable to or greater than ampicillin controls. Gas Chromatography- Mass Spectrometry profiling of these two mushrooms revealed four bioactive compounds: 2,4-di-tert-butylphenol, 3-deoxy-D-mannoic lactone, 4-thiazolecarboxylic acid derivative, and 2-formylamino-3-methylbut-2-enoic acid (unique to G. mbrekobenum). Molecular docking indicated strong binding affinities of these compounds for key bacterial enzymes, including dihydrofolate reductase, enolpyruvyl transferase, phospholipase A2, and penicillin-binding protein 1, which supports the observed antibacterial activity. The findings highlight G. mbrekobenum and L. lactinea as promising sources of antibacterial agents, with activity comparable to standard antibiotics. The integration of molecular identification, chemical profiling, and in silico docking provides a robust framework for further discovery of natural products. Further studies should expand antibacterial screening to more pathogens, determine minimum inhibitory concentration and minimum bactericidal concentration values, and validate compound activity through bioassay-guided fractionation.
Keywords: Antibacterial activity, Ganoderma mbrekobenum, GC-MS, Leiotrametes lactinea, Molecular docking.
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
| Microbial screening for antimicrobial residues in cattle tissues processed at slaughterhouses in Kilosa District, Morogoro, Tanzania |
Research Paper
Microbial screening for antimicrobial residues in cattle tissues processed at slaughterhouses in Kilosa District, Morogoro, Tanzania
Mgonja FR, Mpina MC, Mwega ED, Temba BA, Shabaan M, and Mwangwengwa LM.
J. Life Sci. Biomed., 15(4): , 2025; pii:S225199392500011-15
DOI: https://dx.doi.org/10.54203/jlsb.2025.11
Abstract
Antimicrobial residues refer to drug molecules that remain in the meat and organs of slaughtered animals previously treated with antibiotics, particularly when withdrawal periods are not observed. The inappropriate use of antimicrobial drugs, limited knowledge among livestock stakeholders, and practices such as self-medication contribute to the accumulation of these residues in edible tissues. This study aimed to investigate the presence of antimicrobial residues in cattle tissues using microbial screening methods at slaughterhouses in Kilosa District. A cross-sectional study was conducted in three wards: Kimamba, Dumila, and Kilosa. A total of 90 tissue samples 30 each of muscle, liver, and kidney were collected. The samples were packaged in sterile polystyrene bags and transported to the laboratory at Sokoine University of Agriculture for analysis. Microbial inhibition methods were used to screen for the presence of antimicrobial residues. Data were analyzed using SPSS version 25. The chi-square test was employed to assess differences among tissue types, while Fisher’s exact test was used to evaluate associations. Overall, 28.9% (26/90) of the tissue samples tested positive for antimicrobial residues. Liver tissues showed the highest prevalence at 46.7% (14/30), followed by muscle tissues at 40.0% (12/30). Surprisingly, no antimicrobial residues were detected in kidney samples. The differences in residue occurrence among the tissue types were statistically significant (p = 0.000). Residue occurrence in muscle tissues varied significantly by ward (p = 0.000), indicating a strong geographic influence. However, the distribution of residues in liver tissues did not show a statistically significant variation among wards (p = 0.314), suggesting a more uniform presence. These findings highlight the urgent need to strengthen regulatory frameworks, promote responsible antibiotic use in livestock production, and enhance surveillance systems. Addressing these issues is critical to protecting public health and combating the growing threat of antimicrobial resistance.
Keywords: Microbial screening methods, antimicrobial residues, antimicrobial, Kilosa district
[Full text-PDF-Inpress] [ePub] [Export citation from ePrint] [How to Cite]
Previous issue | Next issue | Archive
![]() This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
Previous issue | Next issue | Archive
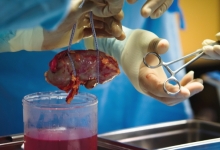
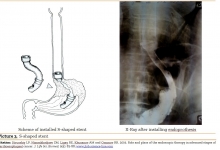
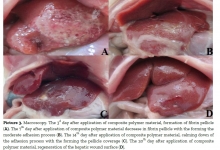
| Comparative analysis of treatment outcomes in chronic pancreatitis complicated by pancreatolithiasis |
Research Paper
Comparative analysis of treatment outcomes in chronic pancreatitis complicated by pancreatolithiasis
Ismailov S, and Mirolimov M.
J. Life Sci. Biomed., 15(2): 42-48, 2025; pii:S225199392500006-15
 DOI: https://dx.doi.org/10.54203/jlsb.2025.6
DOI: https://dx.doi.org/10.54203/jlsb.2025.6
Abstract
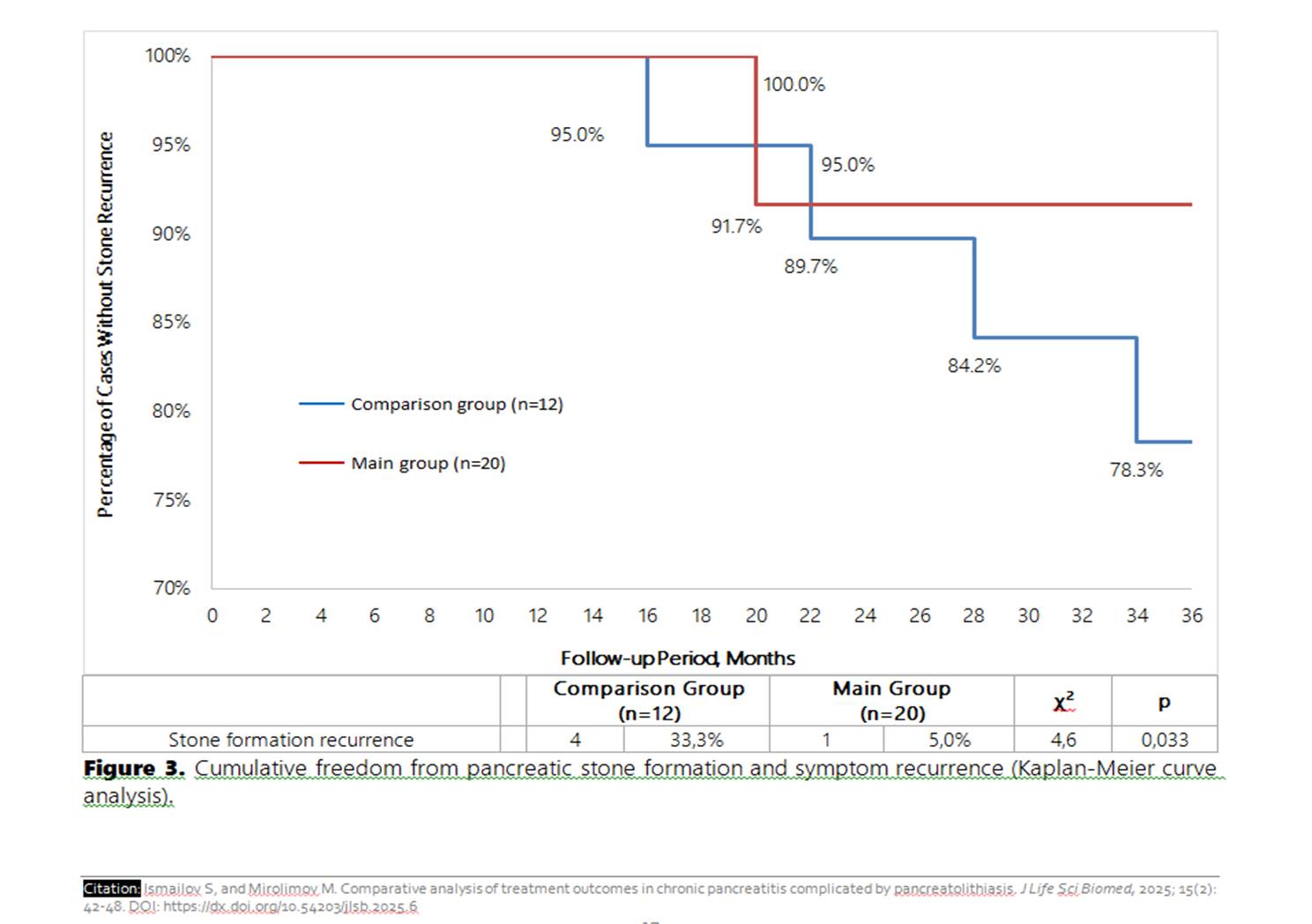
To evaluate the outcomes of a modified Puestow-type pancreaticojejunostomy in patients with chronic pancreatitis complicated by pancreatolithiasis. The study included 32 patients who underwent surgical treatment for pancreatolithiasis. The main group consisted of 20 patients who received the authors' modification of the anastomosis, while the comparison group included 12 patients treated using the standard technique. Parameters assessed included operation duration, mechanical ventilation time, length of hospital stay, complication rates, and recurrence rates. Statistical analysis involved the t-test, χ² test, and Kaplan-Meier survival analysis. The results showed showed a significant reduction in operation time (113.5±6.8 vs. 134.4±5.7 minutes; p=0.025), mechanical ventilation duration (3.0±0.3 vs. 4.25±0.5 hours; p=0.04), and hospital stay (6.3±0.5 vs. 8.4±0.8 days; p=0.033) in the main group. Postoperative complications occurred less frequently, and no reinterventions were required. The recurrence rate of stone formation was reduced from 33.3% in the comparison group to 5.0% in the main group (p=0.033), with cumulative recurrence-free survival of 91.7% versus 78.3%. The modified pancreaticojejunostomy technique provides superior surgical and clinical outcomes in pancreatolithiasis, significantly reducing the risk of recurrence and complications. The study recommended that the modified technique should be considered the preferred surgical option for patients with chronic pancreatitis and pancreatolithiasis.
Keywords: Chronic pancreatitis, pancreatolithiasis, pancreaticojejunostomy, Puestow modification, stone recurrence.
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
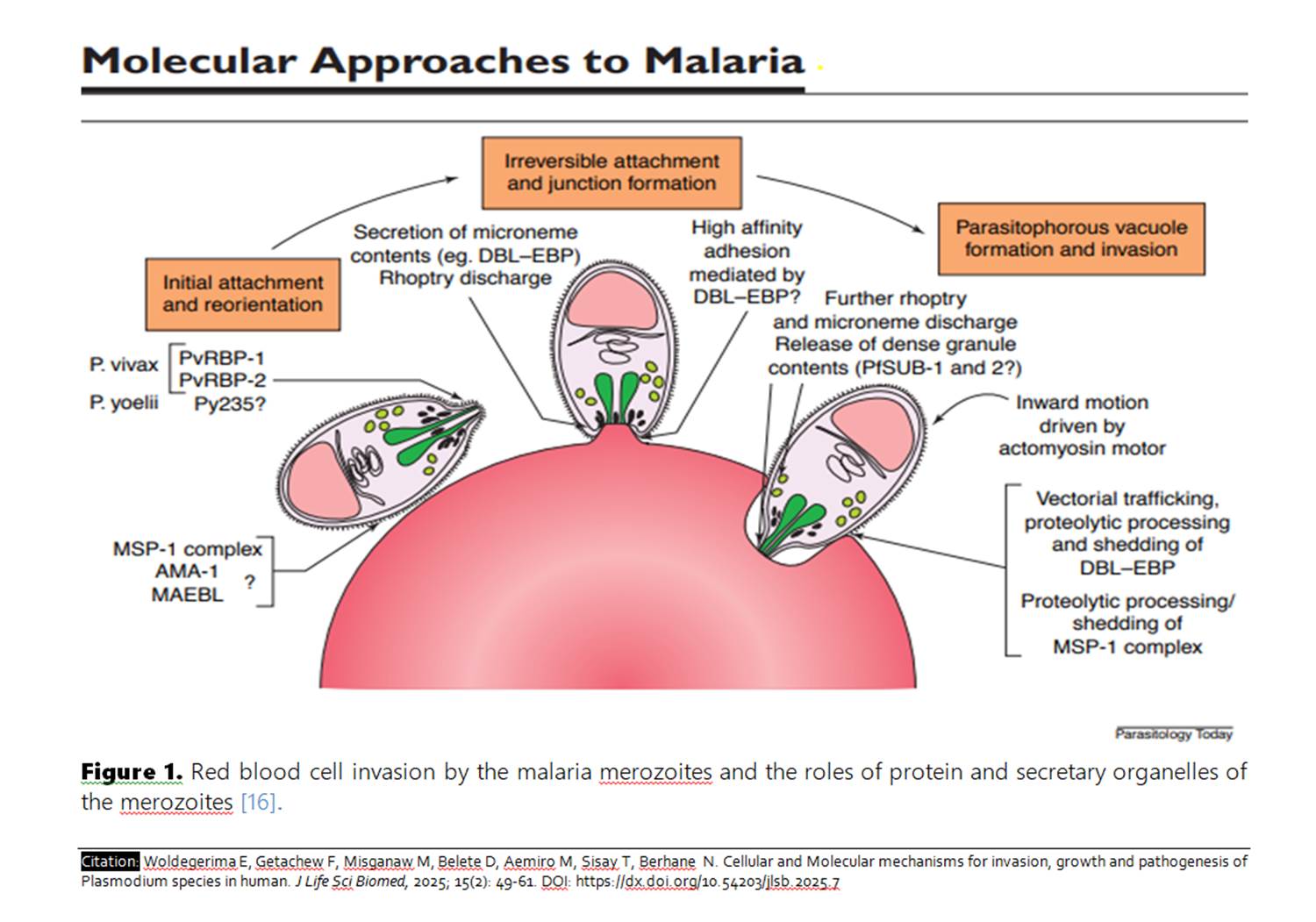
| Cellular and Molecular mechanisms for invasion, growth and pathogenesis of Plasmodium species in human |
Review
Cellular and Molecular mechanisms for invasion, growth and pathogenesis of Plasmodium species in human
Woldegerima E, Getachew F, Misganaw M, Belete D, Aemiro M, Sisay T, Berhane N.
J. Life Sci. Biomed., 15(2): 49-61, 2025; pii:S225199392500007-15
 DOI: https://dx.doi.org/10.54203/jlsb.2025.7
DOI: https://dx.doi.org/10.54203/jlsb.2025.7
Abstract
Malaria is a disease of humans caused by protozoan parasites of the genus Plasmodium with a complex life cycle. Invasion is initiated when merozoites invade circulating erythrocytes. Many proteins, parasite ligands, and host receptors are involved in signaling and erythrocyte membrane fusion. The tight junction and formation of the parasitophorous vacuole membrane must fuse to seal the invasion process. The development of intracellular parasites in conjunction with human evolution has resulted in the establishment of intricate molecular contacts between the parasite and the host cell. These interactions serve the purpose of invading host cells, facilitating migration across different tissues, evading the host immune system, and undergoing intracellular replication. The occurrence of cellular migration and invasion events is crucial for both growth and the development of disease pathogenesis. To review literature written on cellular and molecular mechanisms for invasion, growth, and pathogenesis of Plasmodium species in humans. Literature written on cellular and molecular mechanisms for invasion, growth, and pathogenesis of Plasmodium species in humans was systematically reviewed from 2000–2021 years on Google Scholar sources, Pub Med, and Medline. The key words used to search were erythrocyte, growth, invasion, malaria, and molecular mechanism Pathogenesis, Plasmodium, Red Blood Cell, and Host-parasite Interaction. Malaria is a major health problem caused by protozoan parasites of the genus Plasmodium, whose obligate intracellular life cycle is complex. They use molecular mechanisms to gain access to the host cell and multiply; their apical organelles integrate secretary functions. These secretary organelles, which are proteins in nature, are responsible for successful attachment, reorientation, and invasion of host cells and use Hgb as a nutrient for growth and development. Hgb degradation occurs in an acidic digestive vacuole. During growth, three morphologically distinct phases are observed, and pathogenesis is due to several mechanisms, such as the production of toxins, the sequestration of infected RBC in different organs, the production of inflammatory mediators by the innate and adaptive immune responses, and the hemolysis of RBC. This review was an overview of the molecular and cellular mechanisms for invasion, growth, and pathogenesis of Plasmodium parasites in various aspects of parasite biology and host cell tropism and indicated opportunities for malaria control and the development of an effective vaccine.
Keywords: Cellular, Molecular, invasion, growth, pathogenesis, Plasmodium
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
Previous issue | Next issue | Archive
![]() This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
Previous issue | Next issue | Archive
![]() Volume 15 (3); September 25, 2025
Volume 15 (3); September 25, 2025
| Knowledge, attitude, practice about breast cancer and its screening among female health professionals working at health care facility found in East Gojjam, Ethiopia |
Research Paper
Knowledge, attitude, practice about breast cancer and its screening among female health professionals working at health care facility found in East Gojjam, Ethiopia
Belete D, Tilahun M, Biyazn Z, and Damite W.
J. Life Sci. Biomed., 15(3): 62-70, 2025; pii:S225199392500008-13
DOI: https://dx.doi.org/10.54203/jlsb.2025.8
Abstract
Breast cancer is the leading cause of death among reproductive-age women worldwide and the second leading cause of death among women in Ethiopia. Regular breast self-examination is the most cost-effective method for the early detection of breast cancer. Despite this fact, breast self-examination was low among women in the general population and it was not well documented among health care workers. To explore the knowledge, attitude, and practice about breast cancer and its screening among female health professionals working at public health facilities found in Debre Markos administrative town East Gojjam, Ethiopia. An Institutional-based cross-sectional study design was conducted among female health professions working at public health facilities found in Debre Markos administrative town. A systematic random sampling technique was used from different professionals and a separate sample was taken independently from each. A pre-tested structured questioner was constructed and collects data then Descriptive statistics were used to analyze data by using SPSS version 20. A total of 352 female health professions were included in the study with a response rate of 83.4%. The mean (±SD) age of participants was 24.82(±1.5) years. All of the study participants have heard about breast cancer. 186(52.8%) of them had good, 192 (54.5%) had a positive attitude, 183(52%) have good practice of breast cancer and its screening. 195(55.4%) mentioned that they self-examine their breast at least once in their lifetime, 41 (11.6%) said they had undergone breast examination by clinicians and only five (1.4%) mentioned that they had mammographic screening. Even though the study findings revealed that the majority of study participants had good knowledge of breast cancer, there are still some negative attitudes towards the screenings and practices about breast cancer, there is an alarming increase of breast cancer, therefore the study recommends that, the Ministry of Health and Social Services in collaboration with various stakeholders to develop some strategies on how to address the issues of early detection of breast cancer among female health professions.
Keywords: Knowledge, attitude, practice, Brest cancer
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
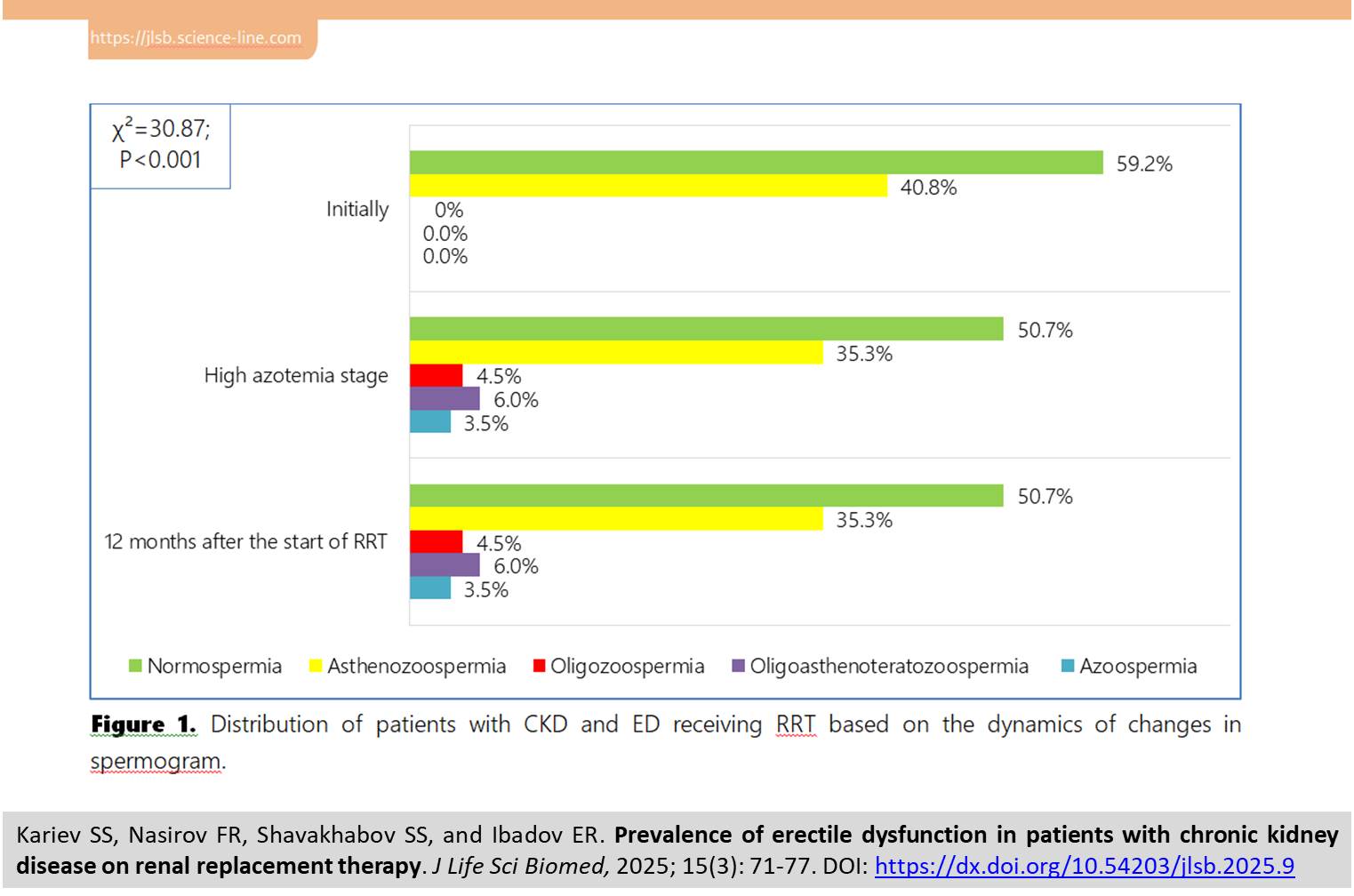
| Prevalence of erectile dysfunction in patients with chronic kidney disease on renal replacement therapy |
Research Paper
Prevalence of erectile dysfunction in patients with chronic kidney disease on renal replacement therapy
Kariev SS, Nasirov FR, Shavakhabov SS, and Ibadov ER.
J. Life Sci. Biomed., 14(3): 71-77, 2025; pii:S225199392400009-13
 DOI: https://dx.doi.org/10.54203/jlsb.2025.9
DOI: https://dx.doi.org/10.54203/jlsb.2025.9
Abstract
The current study aimed to investigate the prevalence of erectile dysfunction (ED) in patients with chronic kidney disease (CKD) on renal replacement therapy (RRT) in the form of hemodialysis (HD). The study included the data of prospective observation of 201 patients on hemodialysis program. The erectile function (EF) was assessed using the International Index of Erectile Function (IIEF‐5) score, doppler sonography of the penile arteries was used to diagnose hemodynamics in the vessels of the penis, the hormonal profile of patients was determined using testosterone, luteinizing hormone, and follicle-stimulating hormone levels, and reproductive function was determined using a spermogram at three stages of the study: initially, high azotemia and 12 months after start of RRT. The results showed that in patients on hemodialysis, ED progression was noted with a decrease in IIEF-5 score from 21.9 to 9.7 points (P<0.001). The average peak systolic velocity in the right cavernous arteries decreased from 6.5±0.1 to 4.8±0.1 cm/s (P<0.001), and also testosterone level, luteinizing hormone, follicle-stimulating hormone, and the frequency of normospermia were significantly decreased (P<0.001), while there was an increase in cases of pathological disorders of spermatogenesis in total up to 14.0% (P<0.001). It is concluded that there is persistence of multifactorial erectile dysfunction caused by vascular, hormonal and structural changes in patients with CKD on RRT in the form of HD.
Keywords: Chronic kidney disease, erectile dysfunction, renal replacement therapy
[Full text-PDF] [ePub] [Export citation from ePrint] [How to Cite]
Previous issue | Next issue | Archive
![]() This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0)
Sustainable Development Goals (UN SDGs)
The Sustainable Development Goals are a call for action by all countries – poor, rich and middle-income – to promote prosperity while protecting the planet. They recognize that ending poverty must go hand-in-hand with strategies that build economic growth and address a range of social needs including education, health, social protection, and job opportunities, while tackling climate change and environmental protection.
Journal of Life Science and Biomedicine (JLSB) is pleased to declare its commitment to the United Nations Sustainable Development Goals (UN SDGs) Publishers Compact. It aims to accelerate progress to achieve SDGs by 2030.
Signatories aspire to develop sustainable practices and act as champions of the SDGs during the Decade of Action (2020-2030), publishing books and journals that will help inform, develop, and inspire action in that direction.
Furthermore, Scienceline Publications, Ltd and its journals like JLSB as a signatory of SDG Publishers Compact commit to:
1. Committing to the SDGs: Stating sustainability policies and targets on our website, including adherence to this Compact; incorporating SDGs and their targets as appropriate.
2. Actively promoting and acquiring content that advocates for themes represented by the SDGs, such as equality, sustainability, justice and safeguarding and strengthening the environment.
3. Annually reporting on progress towards achieving SDGs, sharing data and contribute to benchmarking activities, helping to share best practices and identify gaps that still need to be addressed.
4. Nominating a person who will promote SDG progress, acting as a point of contact and coordinating the SDG themes throughout the organization.
5. Raising awareness and promoting the SDGs among staff to increase awareness of SDG-related policies and goals and encouraging projects that will help achieve the SDGs by 2030.
6. Raising awareness and promoting the SDGs among suppliers, to advocate for SDGs and to collaborate on areas that need innovative actions and solutions.
7. Becoming an advocate to customers and stakeholders by promoting and actively communicating about the SDG agenda through marketing, websites, promotions, and projects.
8. Collaborating across cities, countries, and continents with other signatories and organizations to develop, localize and scale projects that will advance progress on the SDGs individually or through their Publishing Association.
9. Dedicating budget and other resources towards accelerating progress for SDG-dedicated projects and promoting SDG principles.
10. Taking action on at least one SDG goal, either as an individual publisher or through your national publishing association and sharing progress annually.
JLSB endorses the following SDGs except Goal 11:
 |
 |
 |
||
 |
 |
|||
 |
 |
 |
||
 |
Scienceline Publications, Ltd endorses all 17 goals of the SDGs.
To analyze UN SDGs research using Elsevier’s SciVal, please click here.